Using AI to control energy for indoor agriculture
30 September 2024
Published online 19 May 2010
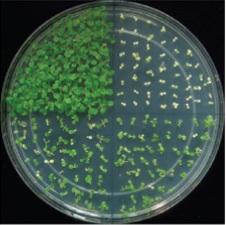
Epigenetic modification of DNA through methylation plays an important role in regulating protein expression in many cells. On activation by 24-nucleotide small-interfering RNAs (siRNAs), the effector protein Argonaute 4 (AGO4) can direct de novo DNA methylation through the methyltransferase DRM2.
However, an international group of researchers, including four from the King Abdullah University for Science and Technology (KAUST) in Saudi Arabia, reported on a new regulator of RNA-directed DNA methylation (RdDM) called RDM1. Mutations in the RDM1 gene led to a marked reduction in methylation in cells isolated from Arabidopsis. The mutations also led to serious impairment of the production of siRNAs. This is similar to the effect of mutations that reduce the production of AGO4, suggesting that both have a downstream role in the effector complex of the RdDM pathway.
The paper suggests that RDM1 binds to single-stranded methyl DNA, which might explain how DNA methylation can control the production and amplification of siRNAs. Through RDM1, the RdDM complex is able to bind to methylated DNA. AGO4 can then induce the formation of the initial siRNAs, more of which are produced through copying. The attachment of AGO4 to the methylated DNA makes it easier for these siRNAs to load onto it, to form functional RdDM complexes.
doi:10.1038/nmiddleeast.2010.149
Stay connected: