Using AI to control energy for indoor agriculture
30 September 2024
Published online 14 September 2014
© Nature Communications
Enlarge image
An international team of researchers, including Arnab Pain from the King Abdullah University of Science and Technology (KAUST), Saudi Arabia, explored the molecular factors that restrict these related parasites to their specific hosts. Their research, published in Nature Communications1 , aims to understand the potential for these parasites, currently limited to apes, to infect humans.
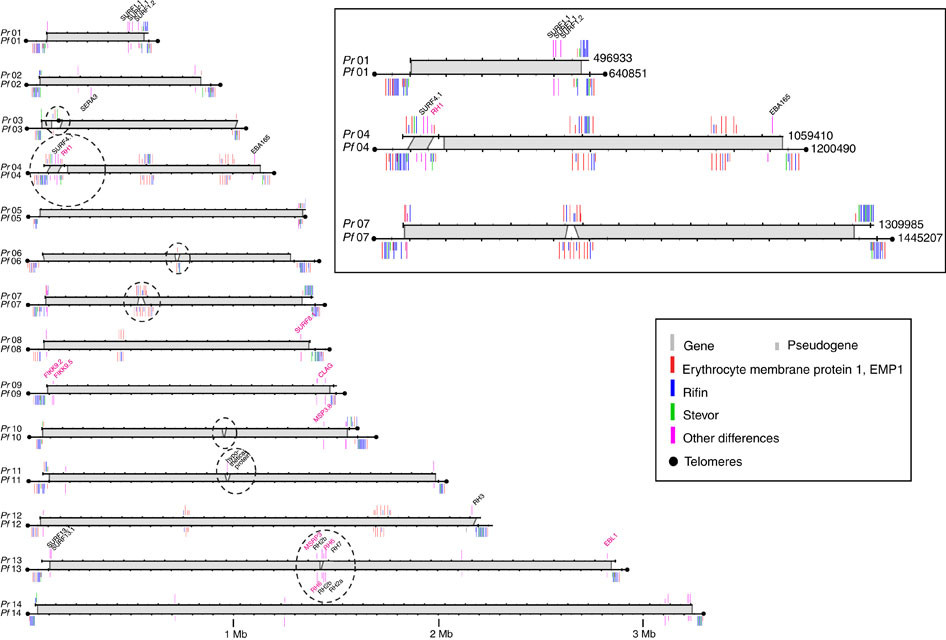
Until now, only the genome of P. falciparum had been sequenced. The researchers sequenced the genome of P. reichenowi and partially sequenced the genome of P. gaboni, both of which infect chimpanzees. They then conducted the first systematic comparison of the three.
They found that P. falciparum and P. reichenowi share almost identical gene content, emphasizing their recent, shared ancestry. The most striking differences were in genes associated with red blood cell invasion. There were also differences in a small group of genes that encode proteins of currently unknown function.
The researchers suggest these variations could be responsible for host specificity, but that still requires further study.
doi:10.1038/nmiddleeast.2014.220
Stay connected: