The state of smart cities in MENA
11 December 2025
Published online 24 September 2014
© Nature Commun
Enlarge image
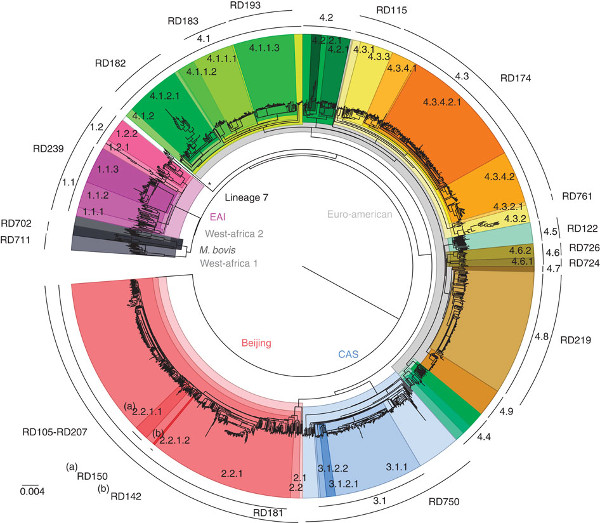
Different strains arise from changes in the sequence of DNA nucleotides in the genomes of the pathogens. Now, a team of researchers from the UK, Portugal and Saudi Arabia, have proposed a system to identify different MTBC strains based on called single-nucleotide polymorphisms (SNPs)1.
By sequencing and comparing the genomes from 1,601 MTBC pathogen populations, they found around 92,000 SNPs including 7,000 that are strain-specific.
From these, they looked for robust SNPs – stable SNPs that are not likely to undergo genetic mutations – and proposed a set of 62 genetic markers, or ‘barcodes’, to genotype common TB strains.
They validated their chosen marker set by testing it on 27 known genomes representing all the MTBC strains. The ability of the set to discriminate different strains was 100%.
Being able to identify the strain causing an infection in a patient may help direct the therapeutic strategy to be undertaken, and is important knowledge for developing vaccines.
doi:10.1038/nmiddleeast.2014.237
Stay connected: