Using AI to control energy for indoor agriculture
30 September 2024
Published online 12 October 2015
Scientists develop a software-based method that can provide insights into molecular organization of the cells.
© Nat. Methods
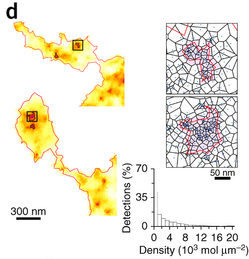
SR-Tesseler subdivides super-resolution microscopic images into polygon-shaped regions centred on localized molecules.
Jean-Baptiste Sibarita and Florian Levet, together with their colleagues from University of Bordeaux, France, and the Ecole Nationale Supérieure de Biotechnologie, Algeria, used simulated and various biological data of proteins labelled with fluorescent markers to test their method.
Simulating single-molecule data of 50-nm-radius clusters with various molecular densities, the scientists found that the method can precisely compute the cluster dimensions and their positions in images.
SR-Tesseler can also automatically analyse microscopic images of both neuronal cell body and molecular cluster of GluA1 subunit, one of the four glutamate acid receptor subunits in neuronal cell cultures. The method successfully studies cell contour, adhesion sites and interaction clusters from live data of fibroblasts expressing integrin beta 3 protein and microtubular structures labelled with fluorescent molecules.
“An important feature of the method is its robustness to noise and molecular density, making it potentially useful for the analysis of molecular organization of various proteins and cell types in two and three dimensions,” says Sibarita.
doi:10.1038/nmiddleeast.2015.182
Stay connected: